Research
Overview
The research interests of the group are broadly centred on protein dynamics and drug design but with a particular interest in membrane proteins. We use and develop a variety of computational tools to improve our understanding of how ligand-binding controls conformational change in proteins. A better understanding of this should enable more rational design of drugs and treatments. Thus much of our work is in collaboration with pharmaceutical companies. We have good local computational resources, but we also make extensive use of ARCHER via the HECBioSim Consortium. We are also founder members of the CCPBioSim - a collaborative computational project for Biomolecular simulation. The following sections provide a little more detail on the areas of current research.
Synaptic Receptors
We have had a long-standing interest in developing and applying computational methods, including docking and molecular dynamics simulations, to receptor proteins and especially ligand-gated ion channels, such as the glutamate receptor or the glycine receptor. In the case of the glycine receptor we were able to predict the binding mode of glycine [1] before it was confirmed by X-ray crystallography.
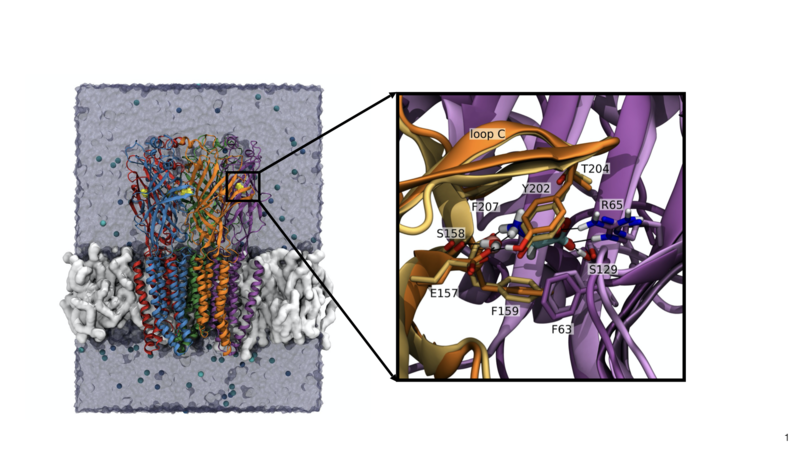
Overlay of our predicted binding mode of glycine (Yu et al. Biochemistry 2014, 53:6041-6051) with X-ray structure (Huang et al. Nature Str. & Mol. Biol., 2016, 24:277-280) solved later
Recently, we were able to resolve a long-standing question in ligand-gated ion channel biology - what does the open state of the glycine receptor look like? Despite its simplicity, it is a non-trivial question to address, because even static structures that might appear to be open from a geometric point of view, the physical environment may still be such that ions and water cannot flow through the gap and thus the channel may still in fact be closed. Furthermore, the open state of such channels is only meta-stable and difficult to study by structural biology techniques. Simulation is an ideal and complementary tool in this case. Through patient and careful molecular dynamics studies we were able to show how the highly conserved leucine residue (at the 9' postion) that is known to act as the main gating point in resting state can adopt a rotameric conformation whereby it swings into its own "mini binding pocket" leading to a pore that is stable and water/ion permeable [2]. This observation has since been confirmed by more recently cryo-EM studies of not only the glycine receptor but other members of the so-called cys-loop receptor family. You can see a movie showing a top-down (ie from the synapse) view of the pore and how these leucines site in perfectly suited "mini binding pockets"
https://www.youtube-nocookie.com/embed/l0_wp95Fk8s
Open state of the Glycine Receptor pore
Collapsed state of the Glycine Receptor pore
Transporters and related proteins
We also have an interest in transporter and transporter-like proteins, in particular those that are likely to be regulated or controlled by pH gradients. For many transporter proteins that rely on a proton gradient to drive the uptake of solutes, the precise mechanistic details of proton coupling remain poorly understood, particularly as structures derived from X-ray crystallography or Cryo-EM can only infer the positions of proteins. MD simulations can provide important information on the role of proton in such cases as exemplified by our recent work on the GkApcT proton-coupled transporter.
Extensive calculations revealed how protonation of a conserved glutamate traps the receptor in an occluded state and that deprotonation is required before the transporter can move to an inward-open state and release substrate in the cell [3]. An example of how the energy landscape of the protein is altered is shown in the figure on the right. The spheres represent the centre of mass that describes the so-callled "collective variable" - this is a convenient, 1-dimensional way of describing the the different conformations that the protein can adopt as it moves between inward-occluded and inward-open states.
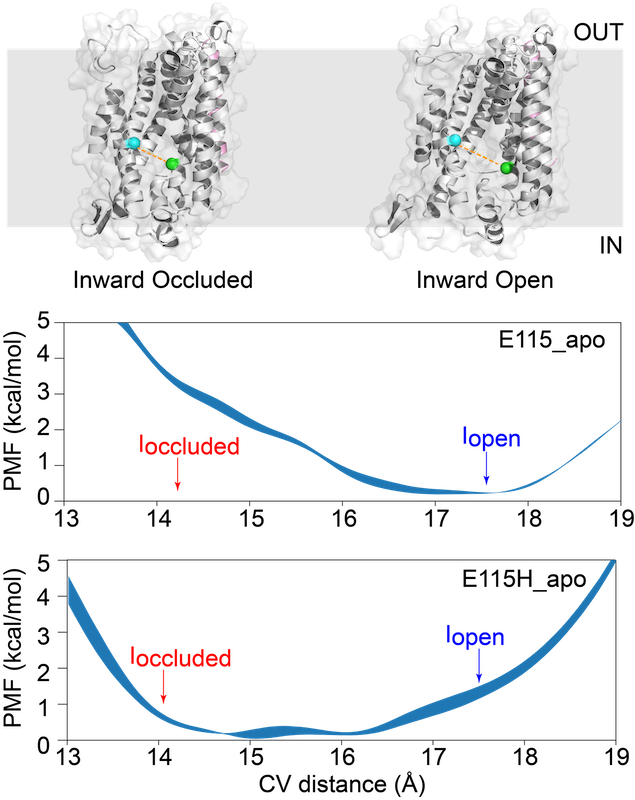
The energy landscape depends on the protonation state of E115. Here, in the apo protein, when no substrate is bound, protonation helps stabilize the inward occluded state. See [3] for more details.
Drug-Design
We are also interested in the more fundamental aspects of drug design. In recent years there has been a lot of discussion about the role of water in drug design. In particular, it has been suggested that by knowing where water molecules bind and being able to predict whether they would be displaceable or not could be used to design compounds that utilize or displace water molecules that are found within binding pockets. One way to predict this is to perform rigorous free energy calculations. However, that is still way too time-consuming to be a realistic tool in high throughput virtual screening. We have developed an approach that is able to predict the positions of water molecules in binding sites and classify them according to whether they will be displaceable or not [4, 5]. More information about that is available from the WaterDock webpage.
Related to this, in the world of virtual screening, scoring functions are often used to provide an indication of how well a compound will bind to a particular protein. We has also show what the maximum error one can expect to see from that type of approach. The results suggest that there is no such thing as a “universal scoring function” and that theoretically you will always be better off using a target-specific scoring function. Most people in the field have recognized this anecdotally, but until now, there was no formal proof [6].
More recent work has focussed on trying to calculate the absolute binding free energy for the binding of small ligands to proteins. Building on the work of many others over a long period of time, we were able to show that the results of these calculations can actually match experimental error [7] and for the first time could also be applied to the much harder problem of selectivity [8]. Moreover with the advent of GPUs technology, the speed of these calculations now means that they can be used to great effectiveness in lead-optimization or small-scale screening activities. Indeed, these approaches have been used in the search for inhibitors of COVID-19 targets.
References
[1] Yu R, Hurdiss E, Greiner T, Lape R, Sivilotti L, Biggin PC (2014) Agonist and antagonist binding in human glycine receptors Biochemistry 53:6041-6051. https://pubs.acs.org/doi/10.1021/bi500815f
[2] Dämgen MA, Biggin PC (2020) A refined open state of the glycine receptor obtained via molecular dynamics simulations Structure 28:1-10. https://doi.org/10.1016/j.str.2019.10.019
[3] Wu Z, Alibay I, Newstead S, Biggin PC (2019) Proton control of transitions in an amino acid transporter Biophys J 17:1342-1351. https://doi.org/10.1016/j.bpj.2019.07.056
[4] Ross GA, Morris GM, Biggin PC (2012) Rapid and accurate prediction and scoring of water molecules in protein binding sites PLoS ONE 7:e32036. https://doi.org/10.1371/journal.pone.0032036
[5] Sridhar A, Ross GA, Biggin PC (2017) Waterdock 2.0: Water placement prediction for Holo-structures with a pymol plugin PLoS ONE 12:e0172743. https://doi.org/10.1371/journal.pone.0172743
[6] Ross GA, Morris GM, Biggin PC (2013) One size does not fit all: The limits of structure-based models in drug discovery J Chem Theor Comput 9:4266-4274. https://pubs.acs.org/doi/10.1021/ct4004228
[7] Aldeghi M, Heifetz A, Bodkin MJ, Knapp S, Biggin PC (2016) Accurate calculation of the absolute free energy of binding for drug molecules Chem Sci 7:207-218. https://doi.org/10.1039/C5SC02678D
[8] Aldeghi M, Heifetz A, Bodkin MJ, Knapp S, Biggin PC (2017) Predictions of ligand selectivity from absolute binding free energy calculations J Am Chem Soc 139:946-957. https://pubs.acs.org/doi/10.1021/jacs.6b11467



